For the last few decades, researchers have routinely used museum specimens to obtain historical genetic information. These data can help to construct phylogenetic trees and track temporal changes in population structure. Specimens have been preserved through drying and liquid preservation for well over a century and, more recently, tissues have been frozen specifically with DNA preservation in mind. Using a variety of approaches, DNA can be extracted and analysed from almost any specimen.
Until very recently, an entire wing of the collection vault has been off limits to genetic analyses – those specimens preserved with the fixative formaldehyde. My team has produced a reliable and robust pipeline for vetting formalin-preserved specimens, extracting and sequencing their DNA.
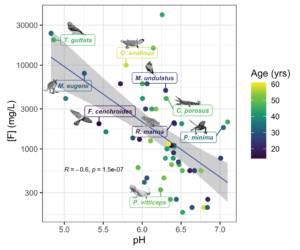 In our 2022 paper published in Molecular Ecology Resources, we tested DNA extraction and library prep methods on a taxonomically diverse set of liquid-preserved specimens collected between 1962 and 2002. From these, we recovered full mitochondrial genomes and up to 3X nuclear genome coverage. We found that hot alkaline lysis coupled with phenol-chloroform extraction out-performs proteinase K digestion approaches. We also show that overall specimen condition (the “ick” factor) driven by post-mortem decomposition prior to fixation is the strongest predictor of extraction success.
In our 2022 paper published in Molecular Ecology Resources, we tested DNA extraction and library prep methods on a taxonomically diverse set of liquid-preserved specimens collected between 1962 and 2002. From these, we recovered full mitochondrial genomes and up to 3X nuclear genome coverage. We found that hot alkaline lysis coupled with phenol-chloroform extraction out-performs proteinase K digestion approaches. We also show that overall specimen condition (the “ick” factor) driven by post-mortem decomposition prior to fixation is the strongest predictor of extraction success.
This approach unlocks genetic data from millions of previously inaccessible specimens!
For a detailed step-by-step protocol, see our recent paper in PLOS ONE and it’s associated bench protocol on protocol.io.
